Reproducibility study of the Fabbri et al. 2017 model of the human sinus node action potential
Abstract¶
The sinoatrial node (SAN) is the natural pacemaker of the mammalian heart. It has been the subject of several mathematical studies, aimed at reproducing its electrical response under normal sinus rhythms, as well as under various conditions.
Such studies were traditionally done using data from rabbit SAN cells. More recently, human SAN cell data have become available, resulting in the publication of a human SAN cell model Fabbri et al., 2017, along with its CellML version.
Here, we used the CellML file provided by the model authors, together with some SED-ML files and Python scripts that we created to reproduce the main results of the aforementioned modelling study.
1Introduction¶
The sinoatrial node (SAN) plays an important role in cardiac function and although it has been extensively studied, some of its intrinsic mechanisms are still open for debate. Most of the experimental data used in SAN modelling have been carried out on animals and on rabbits in particular Lakatta & DiFrancesco, 2009DiFrancesco, 2010Lakatta, 2010Maltsev & Lakatta, 2010Noble et al., 2010Verkerk et al., 2007Himeno et al., 2008DiFrancesco & Noble, 2012Lakatta & Maltsev, 2012Rosen et al., 2012Monfredi et al., 2013Yaniv et al., 2013Yaniv et al., 2015. This has resulted in the development of comprehensive SAN models Wilders, 2007. Yet, this body of work can hardly be transposed to humans.
Human SAN action potentials were first recorded by Drouin (1997), followed by Verkerk et al. (2007) a decade later Verkerk et al., 2007Verkerk et al., 2013. The first human SAN cell model was developed as a proof-of-concept by Seemann et al. (2006). This was followed by the model of Chandler et al. (2009). More recently, Verkerk & Wilders (2015) studied the effect of mutations on human SAN cells, highlighting the need for a human-specific SAN cellular electrophysiology model. Such a model was formulated by Pohl et al. (2016), but its action potential shape does not match that of experimental recordings. Fabbri et al. (2017) addressed this shortcoming by developing their human SAN cell model using available human electrophysiological data.
Fabbri et al. (2017) published a CellML version Cuellar et al., 2003 of their model on the Physiome Model Repository Yu et al., 2011. However, the CellML file on its own is not sufficient to reproduce all predictions presented in the primary paper. Some SED-ML files Waltemath et al., 2011 and Python scripts were therefore created and used with the aforementioned CellML file to reproduce the main results from Fabbri et al. (2017). No modifications were made to the CellML file mathematics or parameters and all the equations and parameters can be found in the original paper.
2Model description¶
Fabbri et al. (2017) developed a human SAN cell model, based on the rabbit SAN cell model of Severi et al. (2012) and on recent electrophysiological data from human SAN cells. The resulting action potential and calcium transient are in agreement with experimentally recorded values. Mutations associated with sinus node dysfunction were also modelled and their effects on pacing rate agree with clinical observations.
The model was developed in Simulink and simulations performed using MATLAB’s ode15s solver Shampine & Reichelt, 1997. Simulations were run until calcium dynamics reached steady‐state. Custom MATLAB (2013a) code was used for automatic optimization and feature extraction. A CellML-encoded version of the model is available at https://
The simulation results presented here were produced using the 2021-07-09 snapshot of OpenCOR Garny & Hunter, 2015 together with various Python scripts that rely on a SED-ML file to configure (the solver to use, the duration of the simulation, the model parameters to track, etc.) and run a given simulation using the model encoded in CellML. Python scripts are also used to generate the figures using Matplotlib Hunter, 2007.
3Model results¶
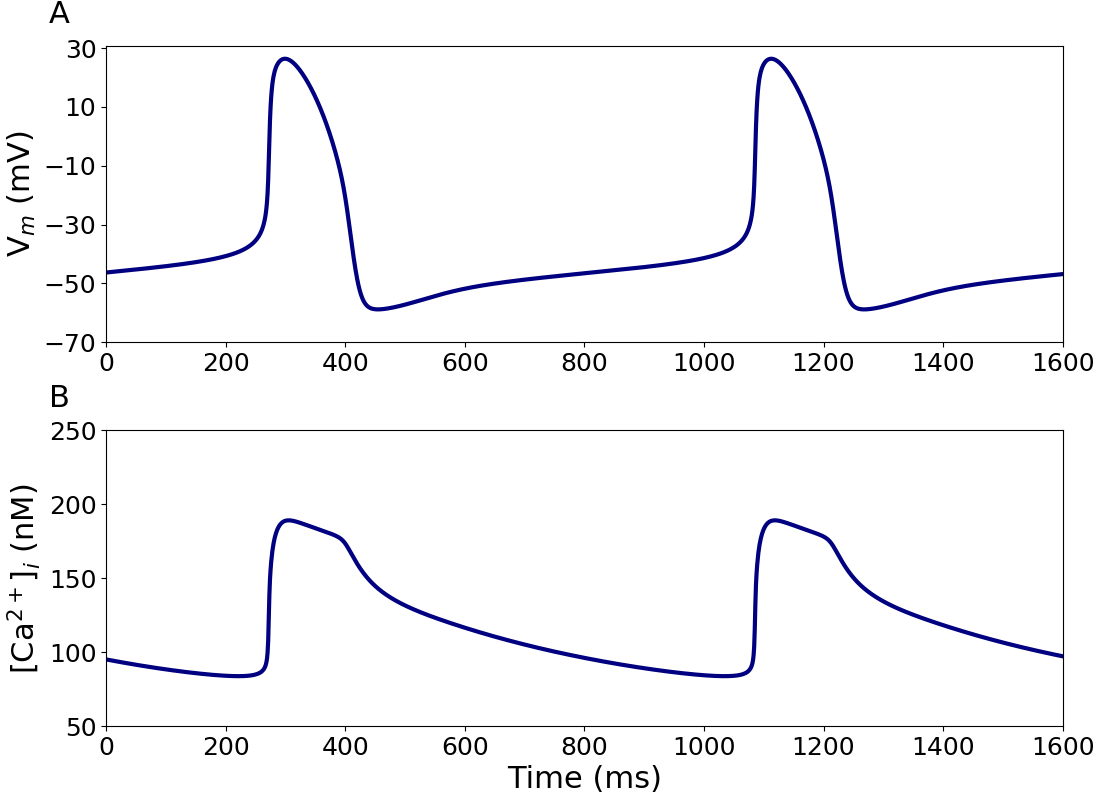
Figure 1:Action potential and intracellular calcium transient of a single human SAN cell.
Simulated action potential (AP; A) and associated calcium transient ([Ca2+]i; B) of a single human SAN cell. This figure can be reproduced using Figure1.py.
Figure 1 shows the action potential (A) and intracellular calcium transient (B), as computed by the model. It reproduces Figure 2 of the primary paper with a cycle length of 814 ms which corresponds to a beating rate of 74 beats min-1. In Figure 2, the time course of the simulated action potential (A) and its underlying components (B-K) are shown, which corresponds to Figure 3 in the primary paper.
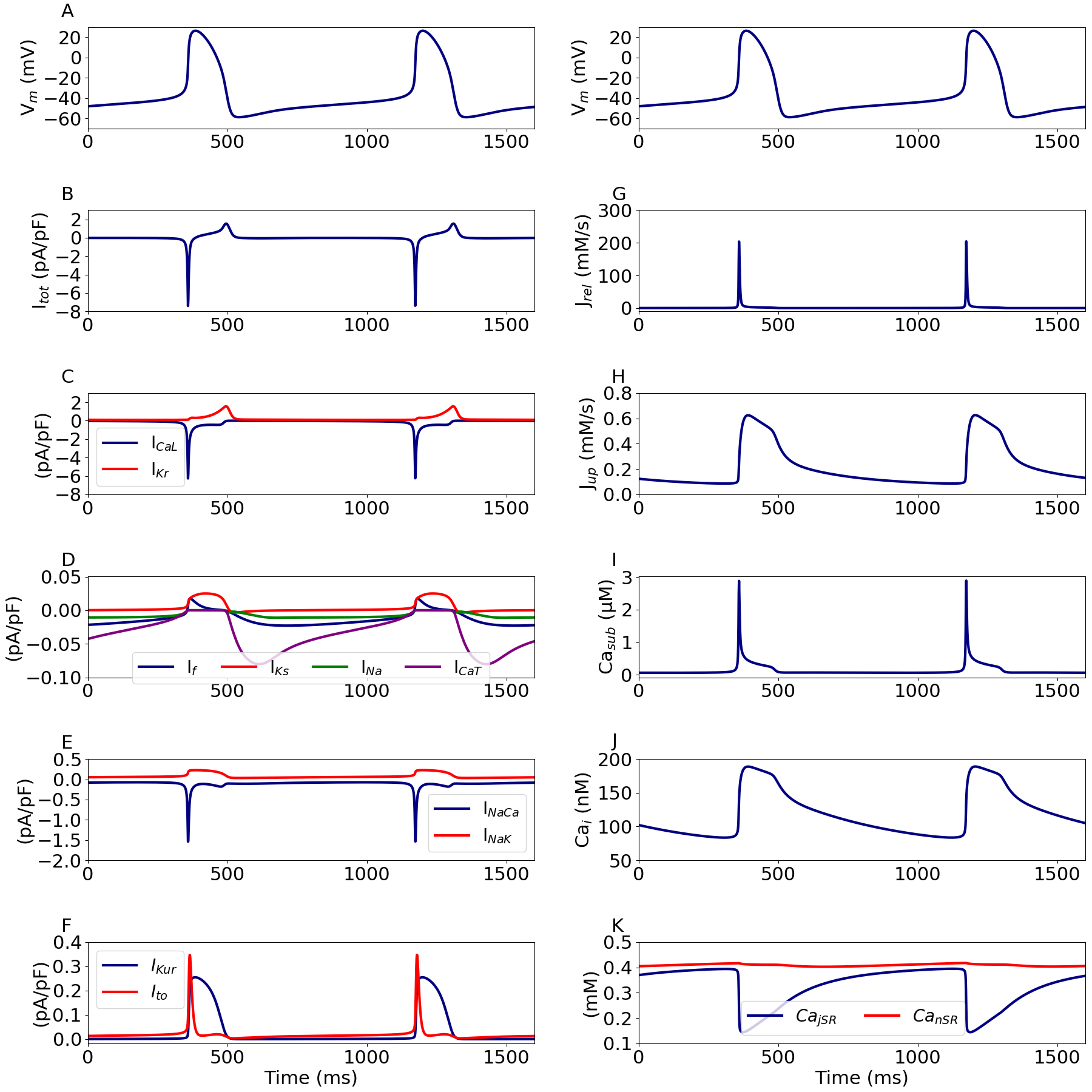
Figure 2:Time course of the simulated action potential and its underlying ionic currents.
Simulated AP (A) and associated currents (B-F), fluxes (G & H), and calcium concentrations (I-K). The top right panel is a copy of panel A and is included for convenience and to match Figure 3 in the primary paper. This figure can be reproduced using Figure2.py.
Figure 3 shows the membrane potential and its associated currents during diastolic depolarization. The resulting behaviour corresponds to that presented in Figure 4 in the primary paper.
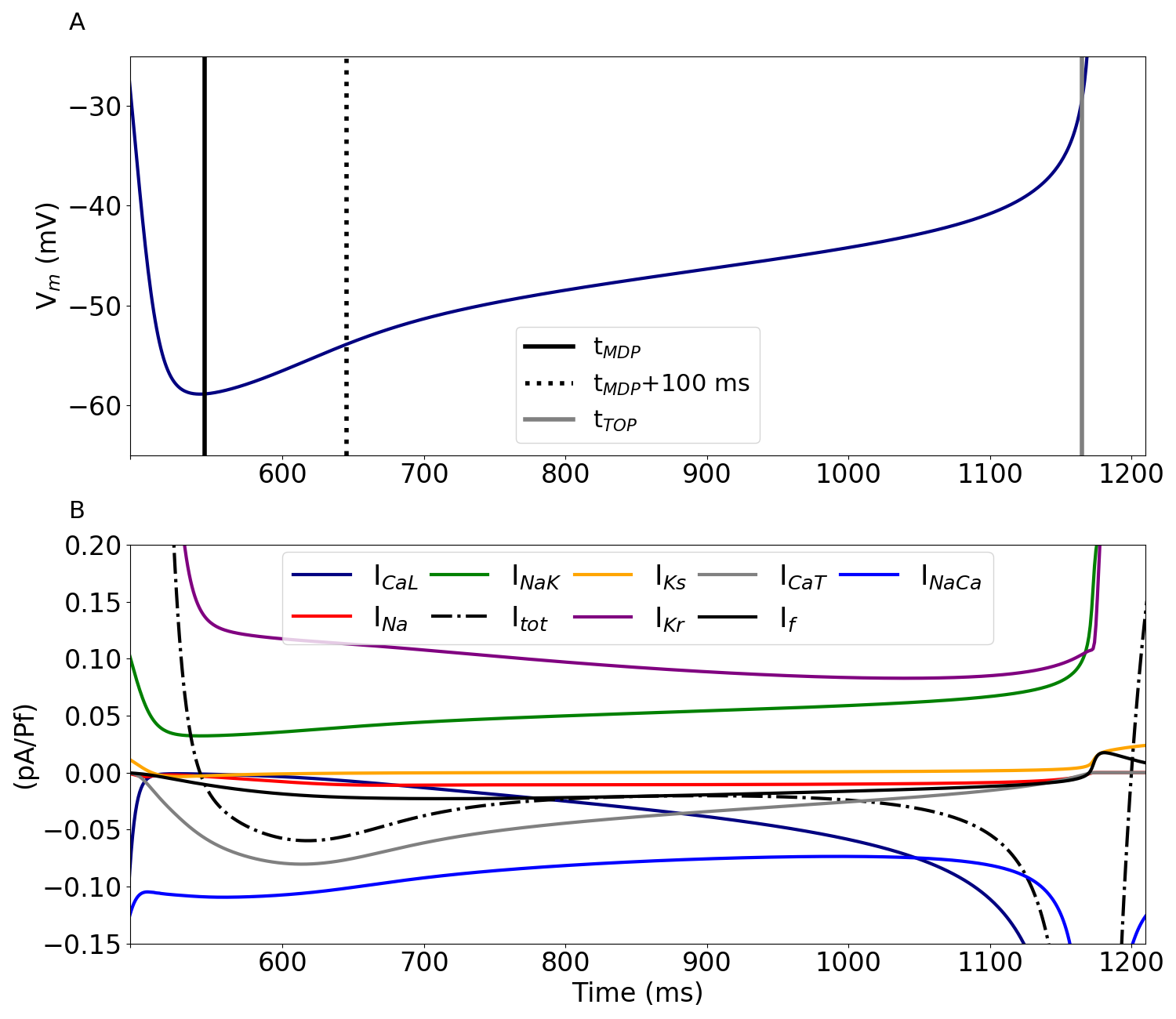
Figure 3:Membrane currents underlying diastolic depolarization.
Simulated AP (A) and associated membrane currents (B). tMDP and tTOP indicate the time at which the membrane potential (Vm) is at its maximum diastolic potential (MDP) and take-off potential, respectively. This figure can be reproduced using Figure3.py.
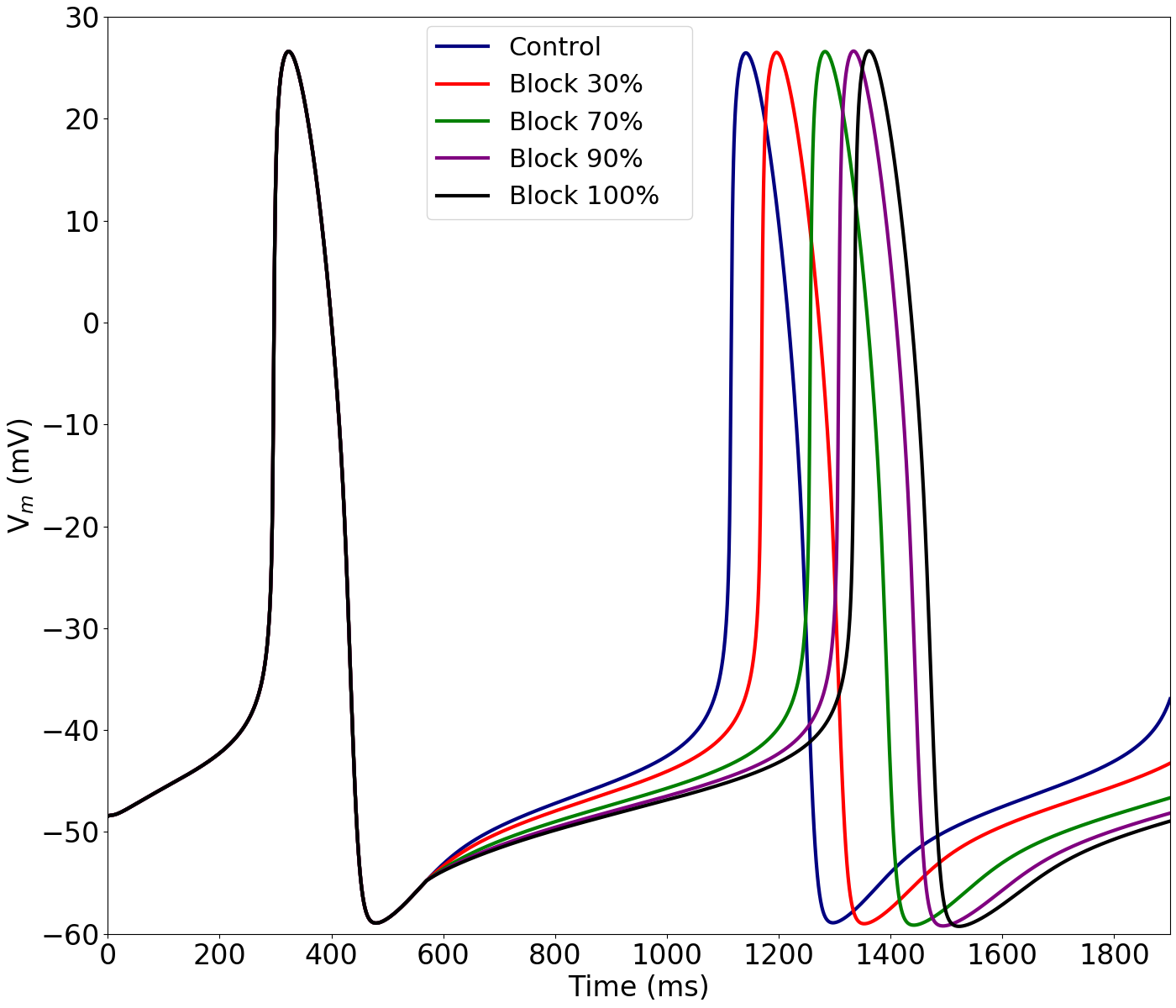
Figure 4:Functional effect of block.
Simulated AP under control conditions (CTRL) and upon 30%, 70%, 90% and full block of the funny current (If). This figure can be reproduced using Figure4.py.
In Figure 4, the model is used to reproduce the results of a progressive block of the funny current, If, to show its effect on the cycle length (CL) and, therefore, on the pacing rate. This corresponds to Figure 7 in the primary paper.
Figure 5 shows the effect of a shift in the activation curve of If on the CL (A), the diastolic depolarization rate over the first 100 ms of diastolic depolarization (DDR100; B), the maximum diastolic potential (MDP; C), and the action potential duration at 90% repolarization (APD90; D). This corresponds to Figure 8 in the primary paper. As reported in Fabbri et al. (2017), little variation in APD90 is observed, albeit with slight differences between the Python implementation presented here and the original MATLAB implementation used in the primary paper.
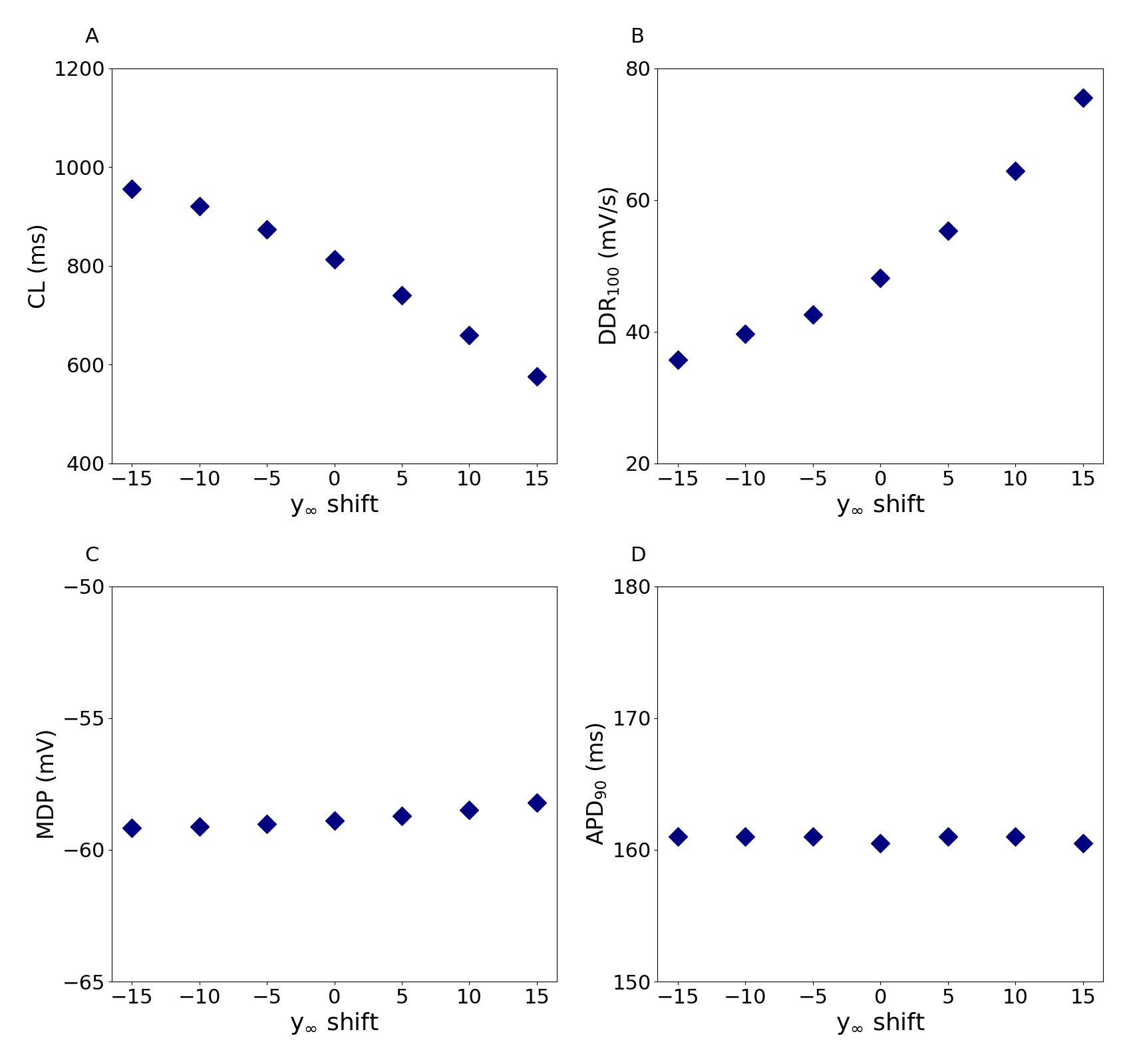
Figure 5:Functional effect of changes in the voltage dependence of activation.
Simulations of the effect of –15 to +15 mV shifts in the voltage dependence of the steady‐state activation curve (y) of If on the cycle length (CL; A), diastolic depolarization rate over the first 100 ms of diastolic depolarization (DDR100; B), MDP (C), and action potential duration at 90% repolarization (APD90; D). This figure can be reproduced using Figure5.py.
The contribution of the sodium-calcium exchanger, INaCa, to the action potential is illustrated in Figure 6, where a progressive block of INaCa was performed. This corresponds to Figure 9 in the primary paper, although the results are slightly different.
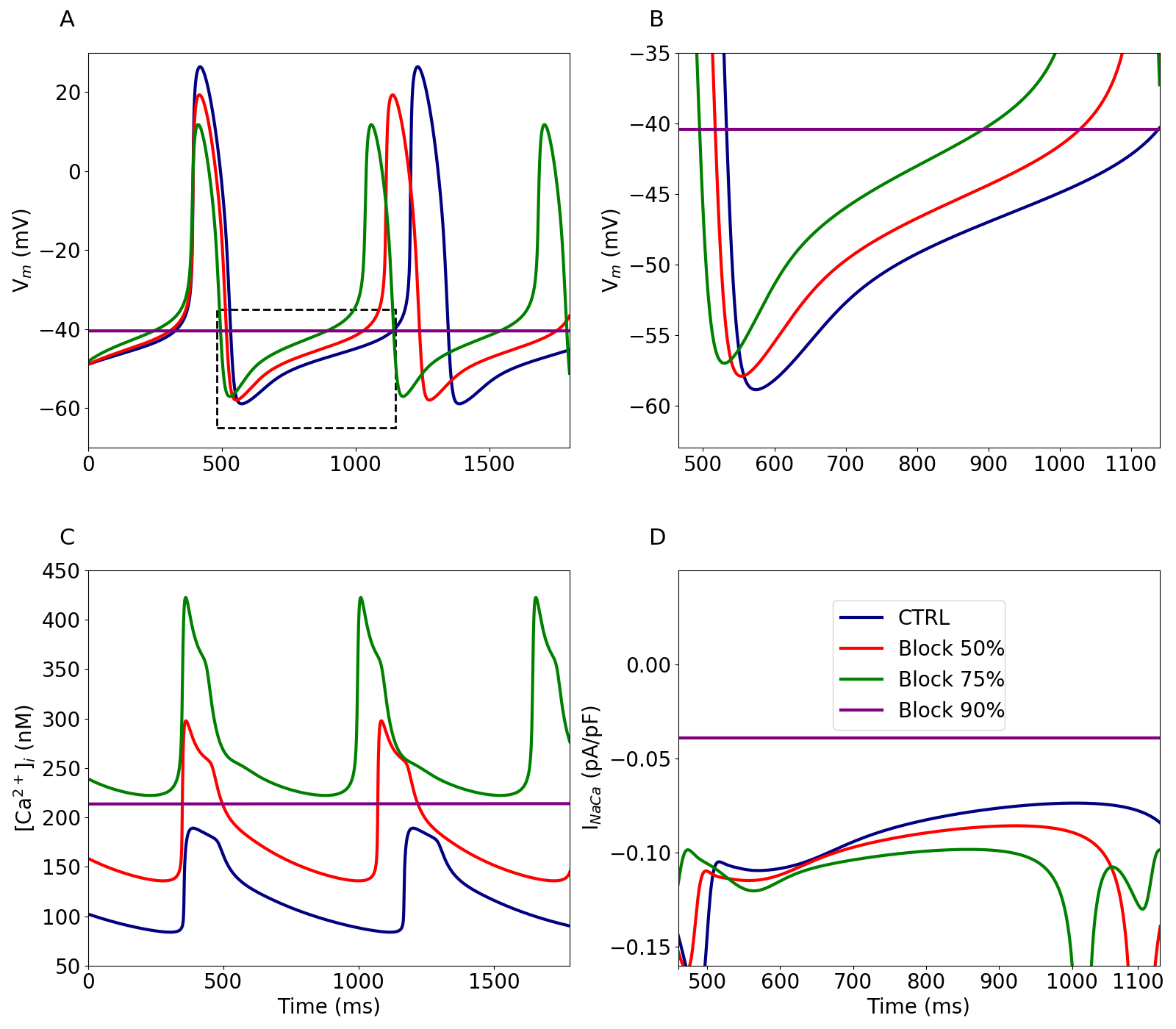
Figure 6:Functional effect of block.
Time course of the simulated AP (A) and associated [Ca2+]i (C) under control conditions (CTRL) and upon 50%, 75% and 90% block of INaCa. Time course of the simulated AP (B) and associated INaCa (D) relative to the dashed box of (A). This figure can be reproduced using Figure6.py.
Figure 7 shows the effect of acetylcholine (ACh) and isoprenaline (Iso) on the membrane potential, its net current, the respective target currents for ACh and Iso, and on the sarco‐endoplasmic reticulum Ca2+‐ATPase (SERCA) pump uptake rate. This corresponds to Figure 10 in the primary paper.
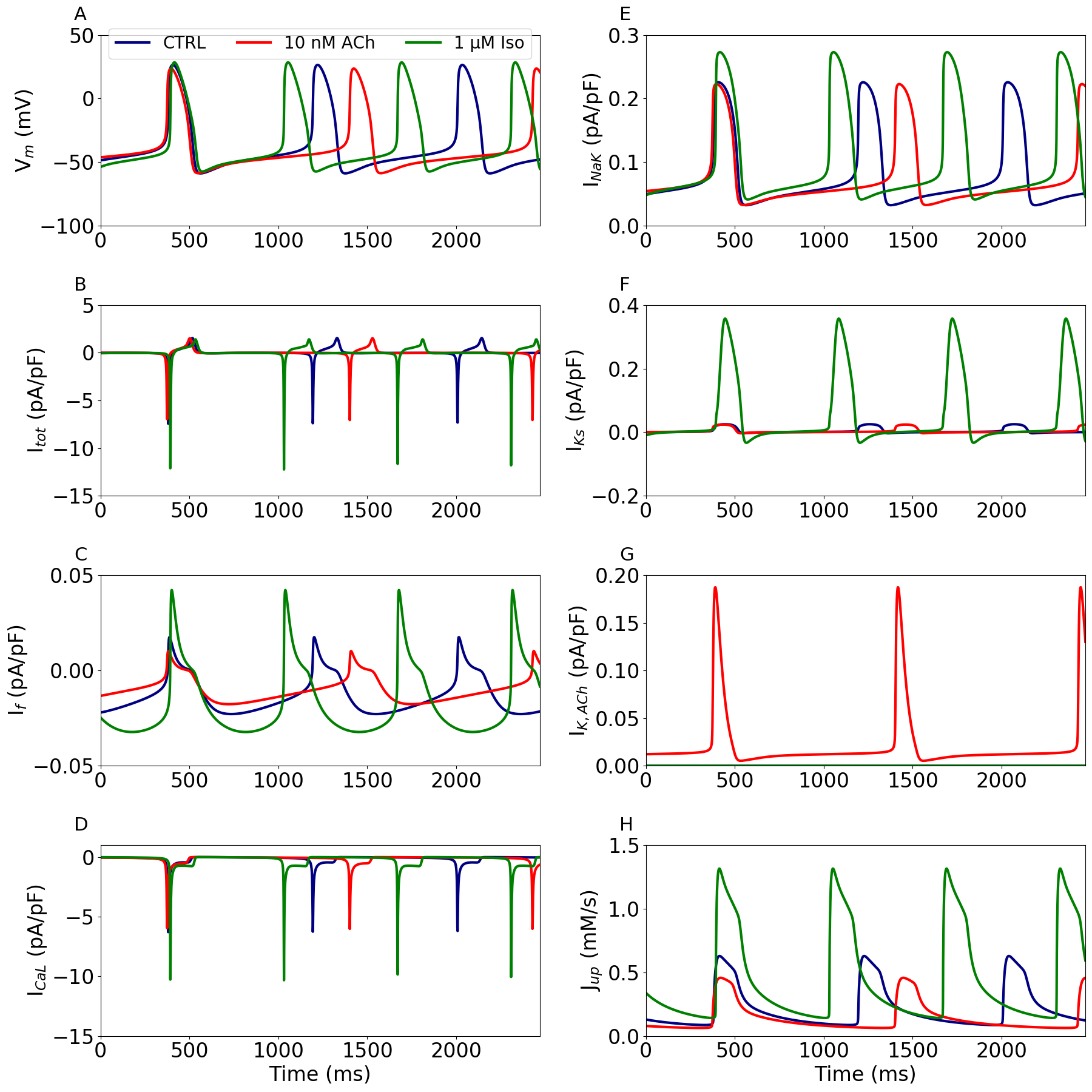
Figure 7:Functional effect of acetylcholine and isoprenaline.
Time course of the membrane potential (A), net current (B), target currents (C–G), and SERCA‐pump uptake rate (H) under control conditions (CTRL) and upon administration of 10 nM acetylcholine (ACh) and 1 M isoprenaline (Iso). The targets for ACh are If, ICaL, IK,ACh and Jup while If, ICaL, INaK, IKs and Jup for Iso. Note the differences in ordinate scales. This figure can be reproduced using Figure7.py.
4Discussion¶
In this manuscript, we used the CellML version of the human SAN cell model that was developed by Fabbri et al. (2017). Most of the main figures in the primary paper were reproducible using the provided CellML code. In some cases, some Python code was needed and can be found at https://
- Fabbri, A., Fantini, M., Wilders, R., & Severi, S. (2017). Computational analysis of the human sinus node action potential: model development and effects of mutations. The Journal of Physiology, 595(7), 2365–2396.
- Lakatta, E. G., & DiFrancesco, D. (2009). What keeps us ticking: a funny current, a calcium clock, or both? Journal of Molecular and Cellular Cardiology, 47(2), 157–170.
- DiFrancesco, D. (2010). The role of the funny current in pacemaker activity. Circulation Research, 106(3), 434–446.
- Lakatta, E. G. (2010). A paradigm shift for the heart’s pacemaker. Heart Rhythm, 7(4), 559–564.
- Maltsev, V. A., & Lakatta, E. G. (2010). Funny current provides a relatively modest contribution to spontaneous beating rate regulation of human and rabbit sinoatrial node cells. Journal of Molecular and Cellular Cardiology, 48(4), 804.
- Noble, D., Noble, P. J., & Fink, M. (2010). Competing oscillators in cardiac pacemaking: historical background. Circulation Research, 106(12), 1791–1797.
- Verkerk, A. O., Wilders, R., van Borren, M. M., Peters, R. J., Broekhuis, E., Lam, K., Coronel, R., de Bakker, J. M., & Tan, H. L. (2007). Pacemaker current (I f) in the human sinoatrial node. European Heart Journal, 28(20), 2472–2478.
- Himeno, Y., Sarai, N., Matsuoka, S., & Noma, A. (2008). Ionic Mechanisms underlying the positive chronotropy induced by β1-adrenergic stimulation in guinea-pig sinoatrial node cells: a simulation study. The Journal of Physiological Sciences, 0801180027–0801180027.
- DiFrancesco, D., & Noble, D. (2012). The funny current has a major pacemaking role in the sinus node. Heart Rhythm, 9(2), 299–301.
- Lakatta, E. G., & Maltsev, V. A. (2012). Rebuttal: What If the shoe doesn’t fit?“The funny current has a major pacemaking role in the sinus node.” Heart Rhythm, 9(3), 459–460.
- Rosen, M. R., Nargeot, J., & Salama, G. (2012). The case for the funny current and the calcium clock. Heart Rhythm, 9(4), 616–618.
- Monfredi, O., Maltsev, V. A., & Lakatta, E. G. (2013). Modern concepts concerning the origin of the heartbeat. Physiology, 28(2), 74–92.
- Yaniv, Y., Sirenko, S., Ziman, B. D., Spurgeon, H. A., Maltsev, V. A., & Lakatta, E. G. (2013). New evidence for coupled clock regulation of the normal automaticity of sinoatrial nodal pacemaker cells: Bradycardic effects of ivabradine are linked to suppression of intracellular Ca2+ cycling. Journal of Molecular and Cellular Cardiology, 62, 80–89.
- Yaniv, Y., Lakatta, E. G., & Maltsev, V. A. (2015). From two competing oscillators to one coupled-clock pacemaker cell system. Frontiers in Physiology, 6, 28.
- Wilders, R. (2007). Computer modelling of the sinoatrial node. Biopacemaking, 121–148.





